The Lichtarge Computational Biology Lab combines novel evolutionary equations with machine learning to solve problems in genomic medicine and molecular engineering.
Evolutionary Trace Algorithm
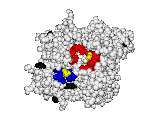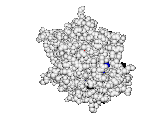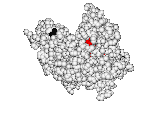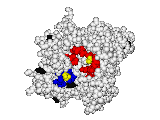
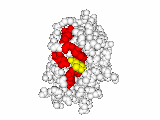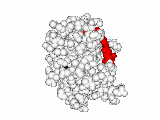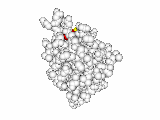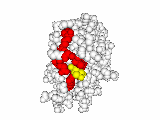
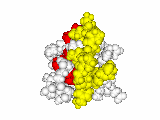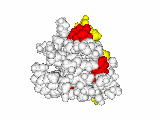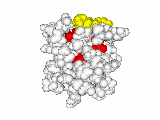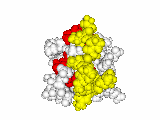
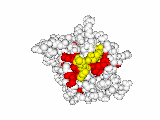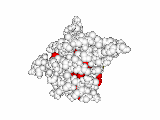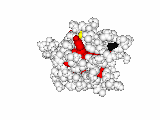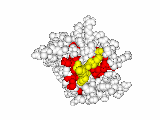
This work began with the goal to identify protein functional surfaces. For this, we sought to compute evolutionarily important sequence positions and introduced the Evolutionary Trace (ET) method. This algorithm was the first to explicitly use phylogenetic divergences to weigh the importance of amino acid variations across species1. Systematic studies of protein structure and function established that the more important positions cluster structurally2 to reveal functional sites and their specificity determinants3. With improved scalability4, these characteristic ET determinants could then be matched across the structural proteome to predict functions5 — even substrates in favorable cases6
G Protein Signaling
A hallmark of ET development is that it was largely motivated by and applied to collaborative studies to elucidate G protein signaling. Discoveries with Bourne, Wensel, Bouvier, Caron and Lefkowitz included: G protein binding sites7; an allosteric switch in Regulators of G protein Signaling proteins8,9; and transmembrane micro-domains for ligand binding, allosteric triggering, and effector coupling in G Protein-Coupled Receptors10. ET also guided separation of function mutations in bioamine receptors, notably decoupling G protein from ß-arrestin effector pathways in vitro11, and, later, in mice12. In other work, mutations targeted in-between the ligand and effector sites rewired a dopamine receptor so that it responded to serotonin, demonstrating allosteric modulation of functional bias13,14.
Together, these ET studies showed how to measure the sensitivity of protein sequence positions to mutations and, as a result, spot most sites that mediate functions and efficiently guides experimental studies that expose and reprogram molecular mechanisms.
Evolutionary Action Equation
Shifting viewpoint, we next reinterpreted ET as the gradient of the fitness landscape. As such, it couples genotype variations with phenotype variations and enables to solve a differential equation for the Evolutionary Action (EA) of mutations on fitness. This EA is computable for most proteins and organisms, and applies across biological scales: In proteins, EA tends to outperform other methods to score the deleterious impact of missense mutations25; in patients, EA correlates with disease morbidity; and in populations, EA explains the distribution of polymorphisms, connecting population genetics to molecular biology15. Clinically, the EA equation also separates head and neck cancer mortality based on p53 mutational severity16, an effect perhaps associated with cisplatin resistance17 and alternative treatment18. EA may also be integrated over patients and pathways25.
Integrative Molecular Biology
Complementary studies focus on machine learning in networks of gene, drug and diseases. Although fine details matter, such as when negative auto-regulatory feedback affects mutational tolerance and evolvability19,20; they are often lacking. This was so in a study that uncovered a new malarial Glutathione-S-Transferase by diffusing information across a gene interaction network spanning nearly 400 species. This new GST may play a role in pathogenesis and therapy as it degrades a toxic byproduct of metabolism and is inhibited by Artesunate—the best current antimalarial agent21. At even poorer resolution, a network that adapted IBM’s WATSON to molecular biology and text-mined the entire PubMed corpus of abstracts, nevertheless identified new p53 kinases among other automated hypotheses 22,23.
Together, these studies suggest that machine learning, text-mining, network analyses and evolutionary equations may soon integrate biological information in light of the genome variations most relevant to a given trait, or disease. This will inform studies of the genotype-phenotype relationship across biology; help design individualized therapies based on a patient’s precise and unique genetic location in the human fitness landscape.
To get a comprehensive view of our lab's contributions, explore the core research publications
To read about our previous research, click here.